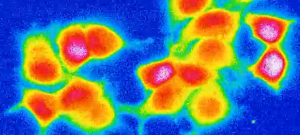
Quantifying information transfer in G-Protein Coupled Receptor signaling
G-Protein Coupled Receptors (GPCRs by their short name) are a very important class of receptors that sit on the cell membrane and inform the cell about events occurring outside the cell. For example, when you smell something it is basically because the neural cells inside your nose have GPCRs that recognize particular chemicals floating in the air. When those chemicals bind to the GPCRs, it triggers a signaling event in the cell, which then processes and relays this information to the brain. Or when you see something, it’s a bunch of GPCRs that sit in the cells of your eyes that gets activated by the light. Because of their centrality GPCRs are the object of many studies, but it was not very clear if single cells could distinguish between a weak and a strong signal or whether they could only distinguish between the presence or the absence of a signal.
In collaboration with the group of Vladimir Katanaev from the Department of Pharmacology and Toxicology, we set out to quantify the amount of information that GPCR signaling can transmit in a single cell. This was made possible by a new experimental setup that they developed, which allowed for the repeated stimulations of single cells with various signal concentrations and the recording of their response in high throughput. Analyzing this data for hundred of cells we could show that cells can reliably distinguish between least 4 different signal strengths. Their response is not only on/off but more like a dimmer with four positions. This has strong implications on the way cells process information from their environment.
Robust gradient formation in the fission yeast
 Fission yeast is a rod-shaped unicellular organism that grows until it reaches about 14 microns and then divides. But wait, how does fission yeast measure it’s own size? It’s a relatively messy process and this project contributed to understanding how yeast can deal with this mess. In some way, we were able to show that yeast can measure its size with a ruler that fluctuates in length! This was done by building a mathematical (more precisely a partial differential equation) model of the measuring tool used by the yeast (a concentration gradient) and theoretically deriving some of its quantitative properties. When we measured those quantities in hundreds of cells, we observed that they closely matched our predictions giving us confidence that the model captures relevant aspect of biological process.
Fission yeast is a rod-shaped unicellular organism that grows until it reaches about 14 microns and then divides. But wait, how does fission yeast measure it’s own size? It’s a relatively messy process and this project contributed to understanding how yeast can deal with this mess. In some way, we were able to show that yeast can measure its size with a ruler that fluctuates in length! This was done by building a mathematical (more precisely a partial differential equation) model of the measuring tool used by the yeast (a concentration gradient) and theoretically deriving some of its quantitative properties. When we measured those quantities in hundreds of cells, we observed that they closely matched our predictions giving us confidence that the model captures relevant aspect of biological process.
This project was a collaboration with Olivier Hachet from the lab of Sophie Martin at UNIL. More information is available here.
This result is interesting because it provides a neat example of how biological systems cope with and even take advantage of their messiness. For a entertaining account of the multiple virtues of messiness, you can read this book.
Regulation of the shade avoidance behavior in Arabidopsis
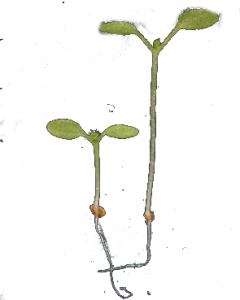
Arabidopsis plants compete again each other for light, which constitutes their primary energy resource. This is especially relevant for seedlings that recently popped out of their seeds, because their ability to capture light is limited by the small size of their two embryonic leaves. Thanks to molecular sensors that react to the (for us invisible) far red light reflected by plants in general, seedlings can « see » that they have neighbors even before they get shaded. This triggers the so-called Shade Avoidance Response, where the seedling will use most of its resources to elongate its stem, in a bid to outgrow its competitors. This piece of research took a dynamical network modeling approach to infer how this behavior is regulated through internal signaling. More precisely we investigated whether the amount of light (and thus energy) available to the plant had an influence on the its behavior. Our results suggest that when light is scarce, the plant saves energy by using weaker internal signal at the cost of a reduced robustness of the behavior.
This project was a collaboration with Séverine Lorrain from the lab of Christian Fankhauser at UNIL. More information is available here.
Sex bias in autism and other neuro-developmental disorders
It has been known for a long time that boys are more often diagnosed with autism than girls. The reason for this were debated in the scientific community, with some arguing that this was the consequence of biases in social norms. By analyzing the genomes of hundreds of autistic persons and their family, we were able to show that males are more sensitive to genetic mutations linked to autism. In other words, females with the same mutations are less likely to have autistic symptoms than males. Why this is so is still unclear, but our results point out at a biological differences between males and females in the susceptibility to autism.
This project was a collaboration with Sébastien Jacquemont from CHUV (now in Montreal ) and the lab of Evan Eichler in the University of Washington. For more details, you can read how the story was covered in The Economist or in La Recherche .
Genome-wide association studies
I was involved in various efforts to find statistical associations between genotypes and heart or blood-related phenotypes in large cohorts.
Machine learning for developmental robotics
 As part of the RobotCub EU project that produced the iCub humanoid robot, I developed bio-inspired machine learning algorithms that enable the robot to learn a representation of its body and peripersonal space in order to control its movement. I also developed algorithms that enables the robot to learn to flexibly execute tasks from human-driven demonstrations.
As part of the RobotCub EU project that produced the iCub humanoid robot, I developed bio-inspired machine learning algorithms that enable the robot to learn a representation of its body and peripersonal space in order to control its movement. I also developed algorithms that enables the robot to learn to flexibly execute tasks from human-driven demonstrations.